Levene’s Test in R Programming
Last Updated :
16 Apr, 2025
Levene's test is an inferential statistic used to assess whether the variances of a variable are equal across two or more groups, especially when the data comes from a non-normal distribution. This test checks the assumption of homoscedasticity (equal variances) before conducting tests like ANOVA.
In this article let's perform Levene’s test in R.
Statistical Hypothesis for Levene's test
A hypothesis is a statement about a given problem, and hypothesis testing is a statistical method used to make decisions based on experimental data. It involves evaluating two mutually exclusive statements about a population to determine which is best supported by the sample data. For Levene's test, the statistical hypotheses areFor Levene's test, the statistical hypotheses are:
Null Hypothesis:
All populations variances are equal
H_0 :\sigma_1^2 = \sigma_2^2 = ...=\sigma_n^2
Alternative Hypothesis:
At least two of them differ
H_1 : \sigma_i^2 \neq \sigma_j^2
The test statistics for Levene’s test are:
W = \frac{\left ( N - k \right )\sum_{i=1}^{K}N_{i} \left ( Z_{i} - Z.. \right )^2}{ \left ( K-1 \right )\sum_{i=1}^{K} \sum_{j=1}^{N_i}\left ( Z_{ij}- Z_i \right )^2}
Levene’s Test in R
R provides a function leveneTest() which is available in the car package that can be used to compute Levene's test.
Syntax: leveneTest(formula, dataset)
1. Levene's test with one independent variable
Consider the R's inbuilt PlantGrowth dataset that gives the dried weight of three groups of ten batches of plants, wherever every group of ten batches got a different treatment. The weight variable gives the weight of the batch and the group variable gives the treatment received either ctrl, trt1, or trt2. To view the random 5 rows of the PlantGrowth dataset use the sample_n() function from the dplyr library.
R
library("dplyr")
print(sample_n(PlantGrowth,5))
Output:
 Sample_Data
Sample_DataWe will test the null hypothesis at 0.05 significance level i.e 95% percentile, considering only one independent variable.
R
install.packages("car")
library(car)
result = leveneTest(weight ~ group, PlantGrowth)
print(result)
Output:
 Output
OutputFrom the result, we can observe that p-value = 0.34 which is greater than our significance level of 0.05. So, we do have not enough evidence to reject the null hypothesis. So the variance across the samples is equal at 0.05 significance level.
2. Levene's test with multiple independent variables:
Let's consider the R's inbuilt ToothGrowth dataset
R
library("dplyr")
print(sample_n(ToothGrowth,5))
Output:
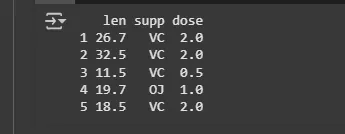 Sample Data
Sample DataIf one wants to do the test with multiple independent variables then the interaction() function must be used to collapse multiple factors into a single variable containing all combinations of the factors.
R
install.packages("car")
library(car)
result = leveneTest(len ~ interaction(supp, dose),
data = ToothGrowth)
print(result)
Output:
 Output
OutputFrom the result, we can observe that p-value = 0.14 which is greater than our significance level of 0.05. So, we do have not enough evidence to reject the null hypothesis. So the variance across the samples is equal at 0.05 significance level.
Other common methods to test for Homoscedasticity of variance across groups include: