This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Snail study points to bright future for AI in biological research

A new James Cook University study using artificial intelligence to analyze the structure of cone snail venom has had mixed results—but points to a bright future for AI in the field of biological research.
Professor Norelle Daly from JCU's Australian Institute of Tropical Health and Medicine, Dr. David Wilson from JCU's Advanced Analytical Center, and Ph.D. student and lead author Tiziano Raffaelli conducted the study, now published in the Journal of Biological Chemistry.
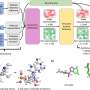
The scientists tested how accurately the AI tool AlphaFold could predict the structure of a specific venom peptide of a cone snail—highly venomous sea snails which can paralyze prey almost instantly and can be a danger to humans.
"The AI successfully predicted the overall structure of the peptide but made errors leading to incorrect predictions about how its stabilizing bonds are formed. While AI software like AlphaFold has made significant strides in predicting larger protein structures, smaller peptides still present challenges," said Professor Daly.
Despite current limitations, Professor Daly believes AI has enormous potential in structural biology, highlighted by the 2024 Nobel Prize in Chemistry awarded to Demis Hassabis and John Jumper for developing AlphaFold.
"AI for structure prediction is incredibly exciting and very promising. I believe it will continue to improve and play a significant role in the field. Although we're not yet at a stage where experimental structural biology can be fully replaced by predictions, studies like this one are crucial for shaping the future of AI predictions."
She said at present, getting structures of peptides is extremely time consuming, costly, and requires specialized equipment and techniques such as crystallography and NMR spectroscopy.
"If we can use AI to accurately predict these structures, it would accelerate the identification and development of novel therapeutics," said Professor Daly.
More information: Tiziano Raffaelli et al, Structural analysis of a U-superfamily conotoxin containing a mini-granulin fold: Insights into key features that distinguish between the ICK and granulin folds, Journal of Biological Chemistry (2024). DOI: 10.1016/j.jbc.2024.107203
Journal information: Journal of Biological Chemistry
Provided by James Cook University